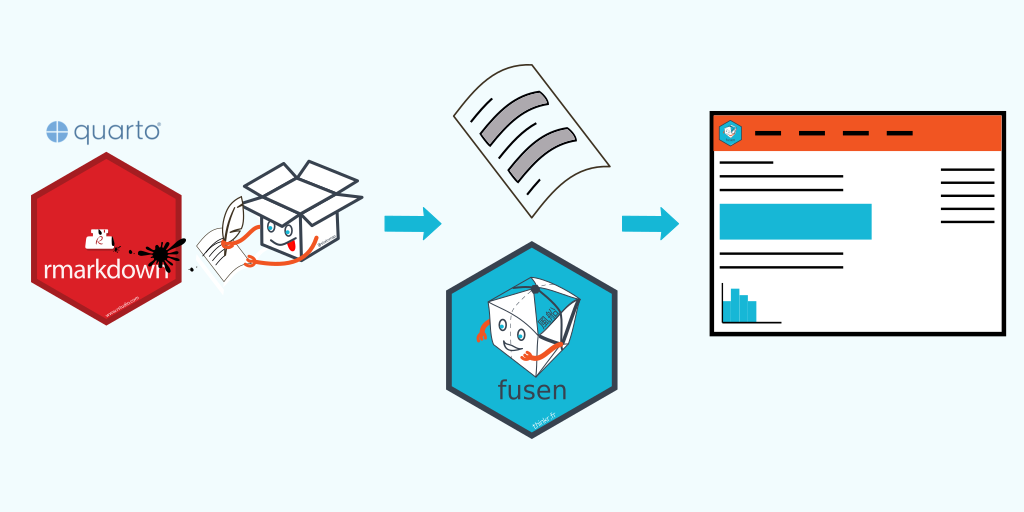
You write R scripts and you would like to share them with other users, in a sustainable and maintainable way. You already write your code in a notebook (Rmarkdown or Quarto) and share its HTML output. You would like to document the chunks, and make this documentation visible in a separate HTML page. Writing a R package seems to be overkill and you would like to experiment good practices for an alternative. Let me show you how {fusen} can help present and share R work following good development practices like doc and test, while starting from a unique place that you already know: the Rmarkdown (or Quarto) notebook.

A fully documented and tested R project, from a notebook to a website in five steps
In the next section, you’ll see the five steps to prepare your project and write your notebook following the template here…
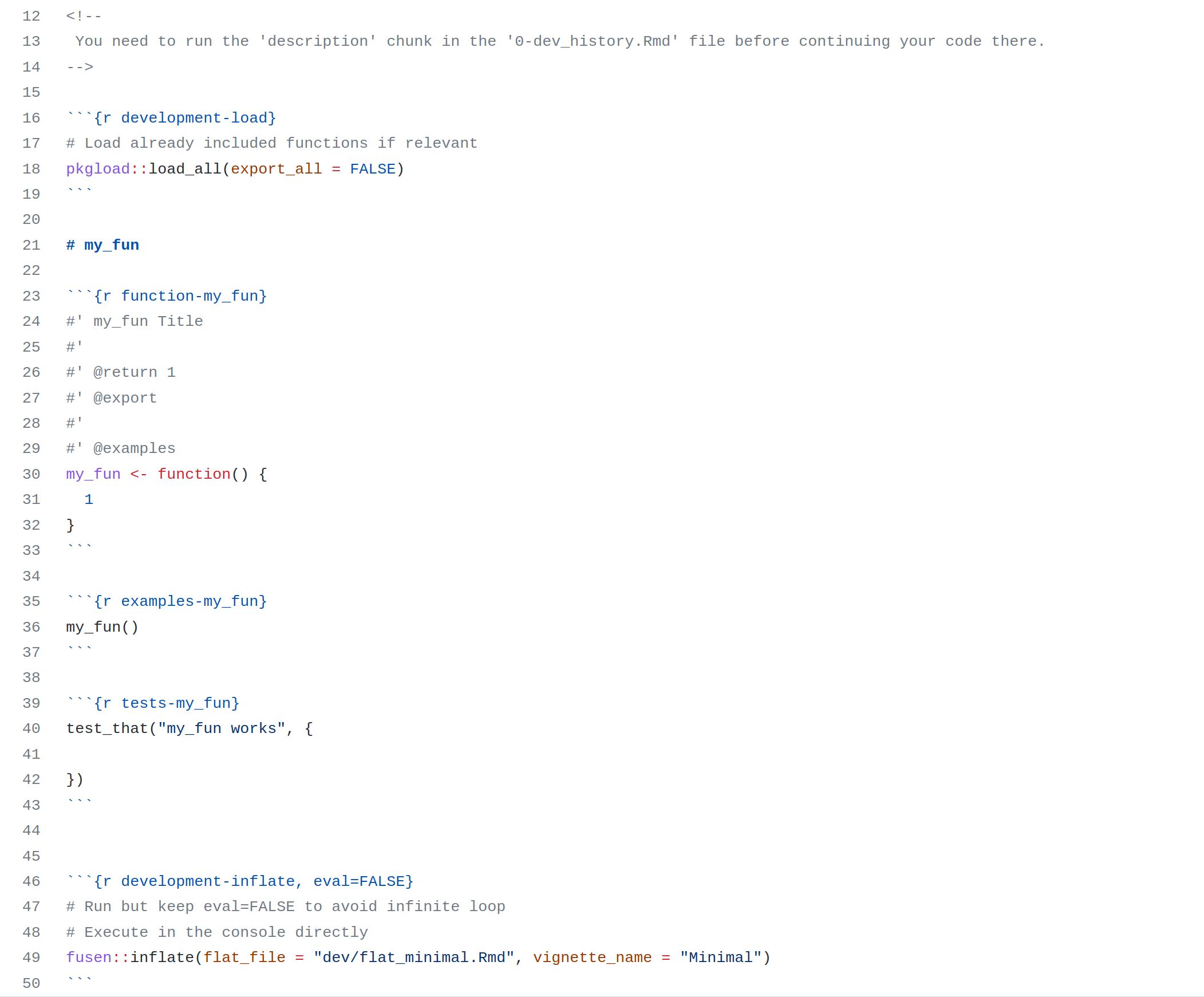
…to create this website there
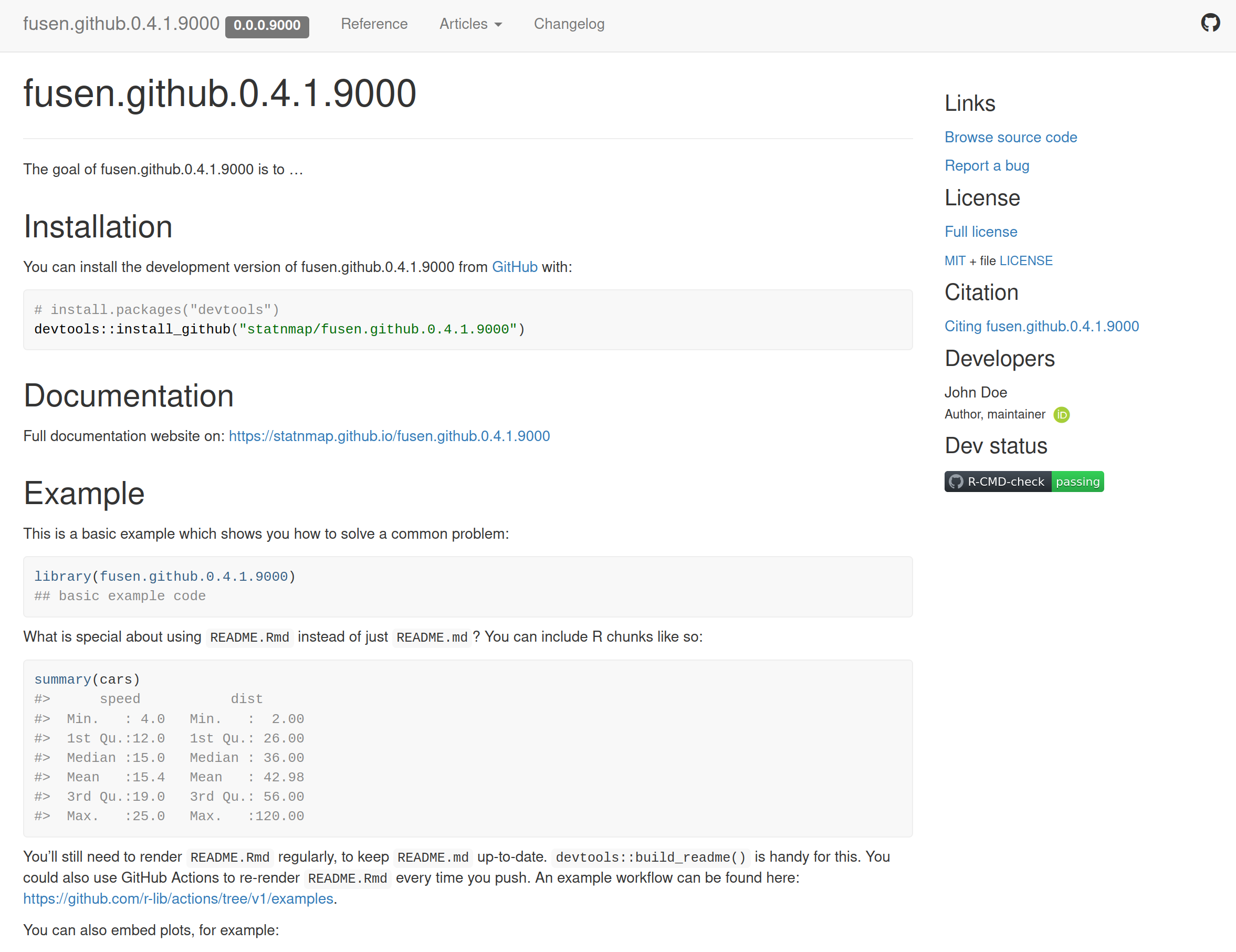
Create your scripts with {fusen} and share them on a GitHub website
You’ll need to install {fusen} from r-universe
You could also install it from CRAN but one of the functionality below is not included on CRAN yet
install.packages("fusen", repos = "https://thinkr-open.r-universe.dev")
1. Create a new project locally
Create a new project with {fusen} using git
- Use
template = "full" if you want explanations on how to fill the sections, or explore {fusen} documentation there: https://thinkr-open.github.io/fusen. Otherwise, template = "minimal" should be enough.
path_project <- "your/path/where/to/start/your/project/project_name"
fusen::create_fusen(path = path_project, template = "minimal", with_git = TRUE, open = TRUE)
2. Describe your project
Follow the “dev/0-dev_history.Rmd” to describe your project
- At least, you will have to fill the
fusen::fill_description() function with your identity as below
# Describe your project
fusen::fill_description(
pkg = here::here(),
fields = list(
Title = "Share Your Project Following Good Development Practices From a Rmarkdown File",
Description = "Use Rmarkdown First method. Strengthen your work with documentation and tests. Everything can be set from a Rmarkdown file in your project.",
`Authors@R` = c(
person("John", "Doe", email = "john@email.me", role = c("aut", "cre"), comment = c(ORCID = "0000-0000-0000-0000"))
)
)
)
# Define License with use_*_license()
usethis::use_mit_license("John Doe")
3. Write your notebook following the skeleton
The first time, you can let the content as is.

4. Inflate the notebook with {fusen}
inflate() the flat template to build the proper structure of the project
- Sections will be transferred in the correct places to get a proper project
- A set of checks will be run to verify that your work is properly documented and tested
fusen::inflate(flat_file = "dev/flat_full.Rmd", vignette_name = "Get started")
5. Share your work on a GitHub website
This is only available on the development version of {fusen} for now
You may need to run usethis::create_github_token(), then gitcreds::gitcreds_set() before, if you do not have a registered GitHub token yet.
Run fusen::init_share_on_github() to create everything needed to set up your website on GitHub. You will be asked multiple times if you want to commit, you can say “yes” each time.
fusen::init_share_on_github()
Carefully read instructions in the console to be able to properly set your GitHub website.
That’s it.
You created a properly documented and tested R project and shared it on a website accessible to everyone.
Every changes pushed to the “main” branch will update the website.
You can see the target website of the project I created for this blog post: https://statnmap.github.io/fusen.github.0.4.1.9000/ and the source repository: https://github.com/statnmap/fusen.github.0.4.1.9000/
Welcome page
 Functions reference
Functions reference
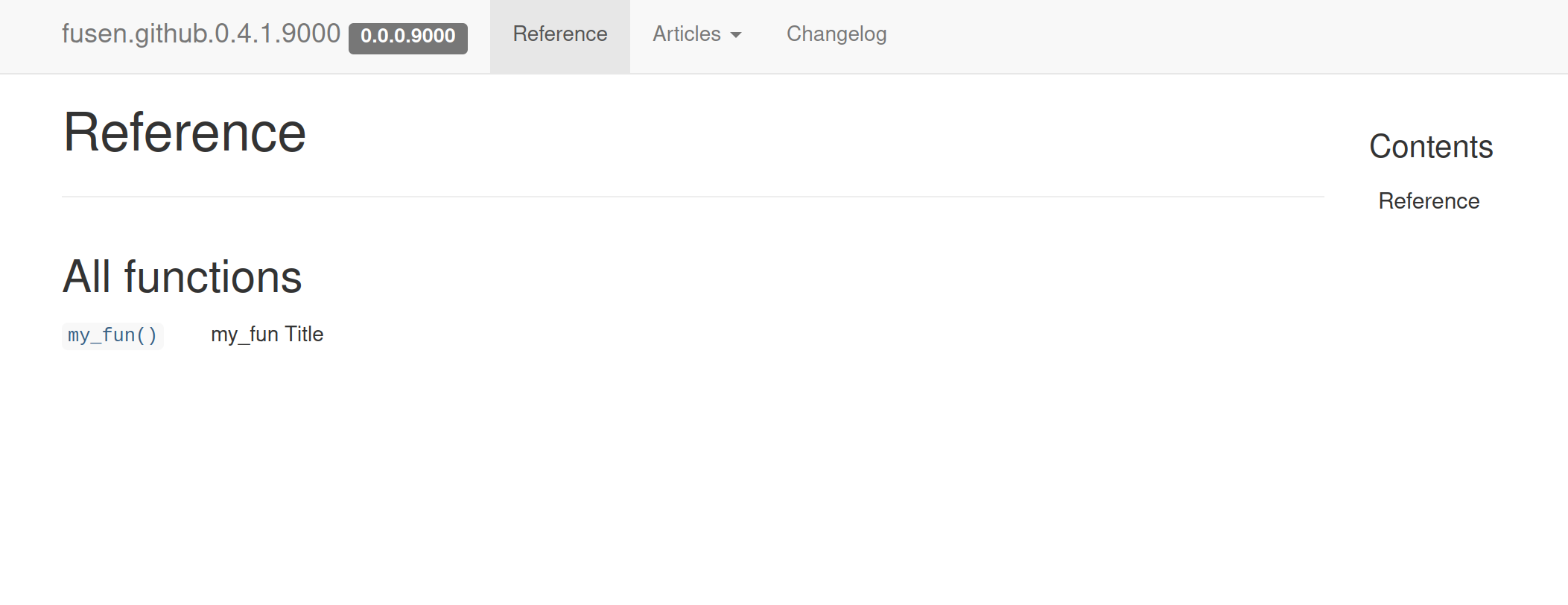 Vignette articles
Vignette articles
 NEWS changelog
NEWS changelog
How to continue the development with the flat template?
Write in plain text what the code is supposed to do before writing any R code.
Then, you will need to factor your code to make it fit in R functions format.
This allows to document and test your code, to make it usable by others, and robust to future changes.
- Start by only capture the whole code in the curly brackets of
myfun <- function() {}
- Then use the function in the
example chunk to create a reproducible example
- When the example is working, you can use it for a unit test in the
test chunk
If you never factorized code or transformed into functions before, I would suggest to follow this presentation: Build reproducible and shareable data analyses using R packages
Use the RStudio Addins “add {fusen} chunks” to add a new set of chunks in your flat file and continue with new features.
Remember to follow the same skeleton as proposed in the flat template to write functions:
- Chunk named
function gets the code and documentation of a function
- Chunk named
example gets the code for examples of using the function. This will be used for function @examples and will be kept for the vignette
- Chunk named
tests gets the code for unit testing
- Chunk named
development gets the code for development purposes, usually only used once like {usethis} functions
Read the different articles in the {fusen} pkgdown website to guide you during the next steps: https://thinkr-open.github.io/fusen/dev/index.html
Yes, I must admit that you created a R package. And its corresponding website with {pkgdown}.
Now, the most difficult part for you is to find a proper hex logo. {fusen} cannot help with that, but {hexmake} can: https://connect.thinkr.fr/hexmake/

How to share my analysis if the flat file presents the documentation of my functions but not my analysis itself?
Indeed, the flat file is not exactly the notebook you want to share. What you want is a report presenting your data. The flat file, along with the website, is more the documentation on how to use the functions you created to analyse your data and the results of the analysis.
This makes the whole work reproducible for other similar analyses. And that’s the whole point! Sharing your work.
In the presentation Build reproducible and shareable data analyses using R packages, there is a specific chapter about What about data analyses in a package?.
You will see that I propose two possible ways to build your analysis reports:
- Classical package: Use external projects to use your package functions in Rmd/Quarto reports. This is probably what you already do now. However, with {fusen}, you can write your R functions inside your package realised with {fusen}, so that they are documented, tested and shared as a website for your future work and for the community.
- Compendium-like: Include the Rmd/Quarto reports inside the package development project, in a specific folder.
In both cases, you can think of your flat template to follow the same steps as in your report (e.g. data preparation, exploration, model output…). So that the vignette of your package, resulting from inflating your template, is your skeleton to present your analysis. This also means that for a new analysis, you can copy the source vignette as a start.
Note: I followed the switch to Mastodon, you can follow me on @statnmap@mastodon.social
Citation:
For attribution, please cite this work as:
Rochette Sébastien. (2022, Oct. 28). "Share your R work following good dev practices from a single notebook". Retrieved from https://statnmap.com/2022-10-28-share-your-r-work-following-good-dev-practices-from-a-single-notebook/.
BibTex citation:
@misc{Roche2022Share,
author = {Rochette Sébastien},
title = {Share your R work following good dev practices from a single notebook},
url = {https://statnmap.com/2022-10-28-share-your-r-work-following-good-dev-practices-from-a-single-notebook/},
year = {2022}
}





 Vignette articles
Vignette articles NEWS changelog
NEWS changelog